
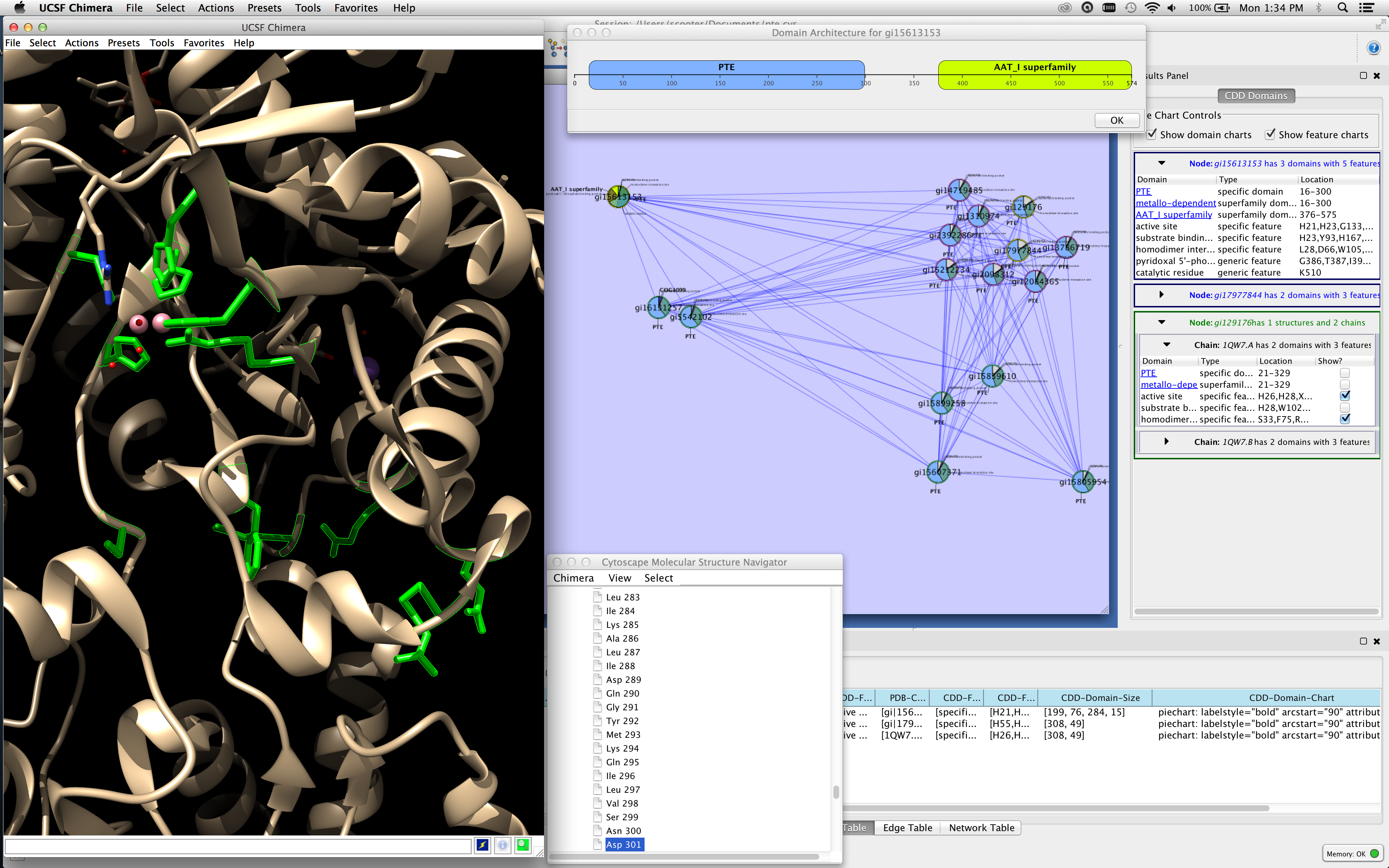
In this blog post, I will try to explain and demonstrate the main features of Cytoscape through a sample visualization of randomly selected Istanbul porters and their guarantors listed in Istanbul Court Record no.24. With the Apps from Cytoscape App Store and other softwares such as R and NetworkX, you can experience advanced features and make sophisticated visualization and analysis. Besides social networks and interpersonal relationships, social scientists can use Cytoscape to visualize and analyze interactions from tables, forms, and the web. The official Cytoscape App page for ANIMO is here.
Cytoscape linux mac os#
When it was initially released in July 2002, it was originally designed for biological studies but then turned into a general platform for different disciplines such as social sciences. ANIMO is available for Windows, Mac OS and GNU/Linux operating systems. The tool also provides numerous ways to annotate your files, and wraps all of its features in a neatly organized interface. For Linux systems, Oracle Java 7 has caused Cytoscape crashes on some platforms.
Cytoscape linux software#
In this respect, Cytoscape is a useful open-source software platform for network analysis and visualization. Cytoscape is an advanced application designed for biology students, researchers and enthusiasts that want a reliable way to view and analyze molecular interaction networks and biological pathways. Keywords: Cytoscape, Interactive Network Visualization, Network Analysis. 1,2.To enable rapid prototyping and release of new methods, Cytoscape is implemented as an open. Network analysis can be applied to various disciplines to depict relational networks and associations among variables such as a group of people, objects, geographical places, and even relationships through nodes, lines, and edges. forms for analyzing these networks is Cytoscape, a general-purpose and freely available software platform for integration, visualization and statistical modeling of molecular networks together with other systems-level data. This repository contains top-level pom file and utility script for building Cytoscape core distribution. Make sure you have Cytoscape installed on your laptop: Cytoscape can be downloaded from. Cytoscape is a fairly complex application and its core distribution has multiple repositories for managing its code. S,-script Execute commands from script file.Network analysis is a method that is consisted of a set of techniques enabling researchers to visualize and analyze the relations among diverse actors and entities. For the computer exercise we will be using Cytoscape for. Other layouts, which are large or less frequently used, are left as external extensions. The builtin layouts are commonly used and they are small in file size. V,-vizmap Load vizmap properties file (Cytoscape VizMap
s,-session Load a cytoscape session (.cys) file. Other parameters of the job, such as the maximum wall clock time, maximum memory, and the job name need to be modified appropriately as well.ĭocumentation module load Cytoscape/3.7.2 Where need to be replaced by the options (command and arguments) you want to use. To run as a batch job, you might have to create a script file with the cytoscape execution commands.Įxample of a job submission script sub.sh to run a batch job (note: this is not the cytoscape execution command script):
A lot of Apps are available for various kinds of problem domains, including bioinformatics, social network analysis, and semantic web.
Cytoscape linux download#
To run this program in an interactive session using the graphical front end: Start an X terminal (Mac), X window (Windows) at the local computer first, then Download 3.9.1 Analyze Your Genes With NDEx iQuery Cytoscape is an open source software platform for visualizing complex networks and integrating these with any type of attribute data. Please refer to Run interactive Jobs and Run X window Jobs. Cytoscape is an open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with. To use version 3.7.2, please first load the module with
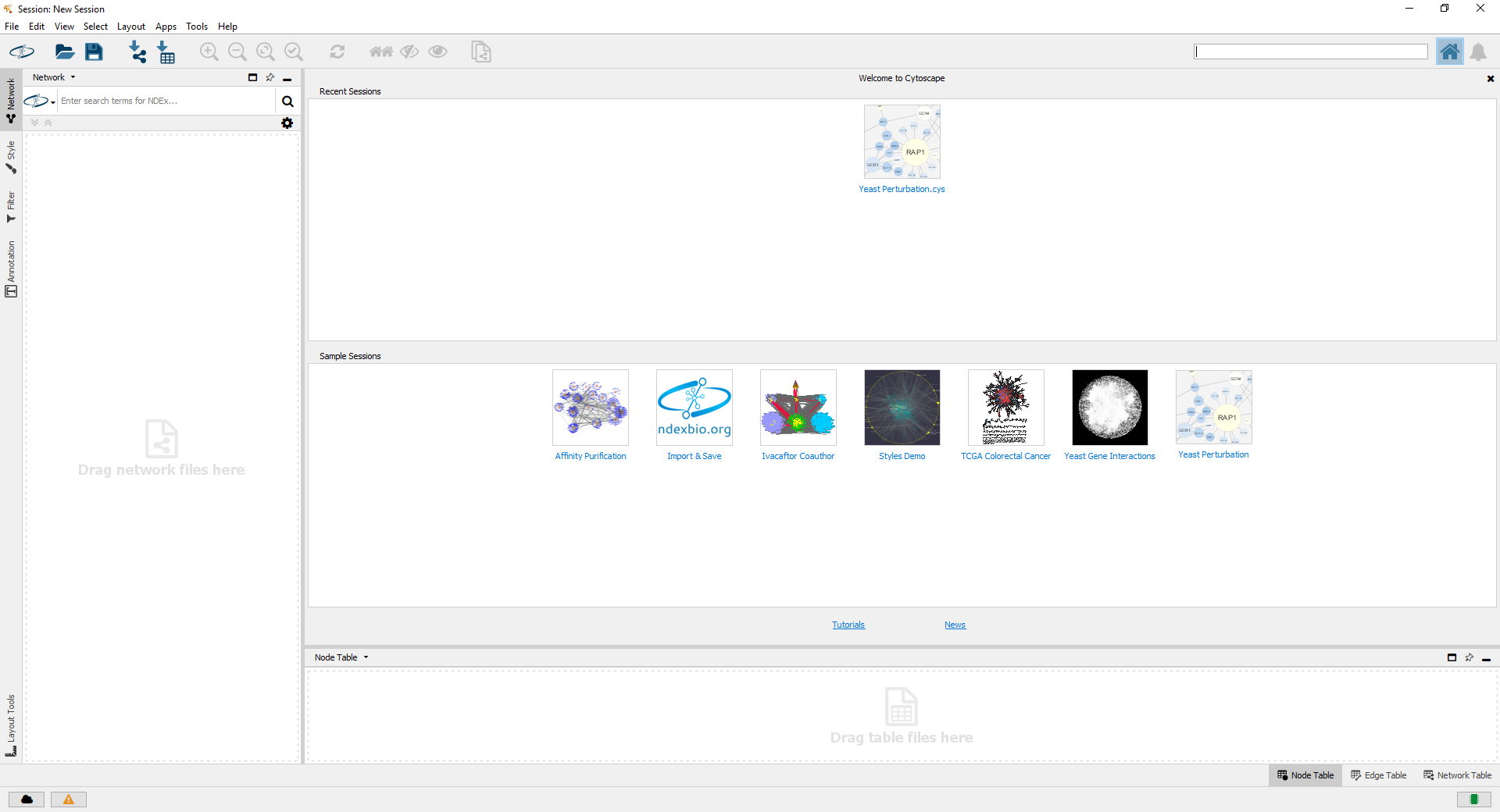
"Cytoscape is an open source software tool for integrating, visualizing, and analyzing data in the context of networks." Cytoscape is available as a platform-independent open-source Java application, released under the terms of the LGPL.


 0 kommentar(er)
0 kommentar(er)
